K-means 클러스터링이란?
주어진 데이터를 K개의 그룹으로 나누는 비지도 학습 알고리즘이다. 이 알고리즘은 군집(클러스터)의 중심을 반복적으로 업데이트하여, 각 데이터 포인트를 가장 가까운 중심(센트로이드)에 할당하는 방식으로 동작한다.
동작방식
- K개의 초기 중심(centroid)을 무작위로 선택한다.
- 각 데이터 포인트를 가장 가까운 중심에 할당한다.
- 각 클러스터의 중심을 다시 계산한다.
- 중심이 더 이상 변하지 않거나, 지정된 반복 횟수에 도달할 때까지 2~3 단계를 반복한다.

목표
K-means 클러스터링을 구현한다.
제약조건
- 두 번의 반복 동안 모든 중심점(centroids)의 위치가 1 * 10^-5 이하로 변할 경우 수렴했다고 간주한다.
- K-means 클러스터링과 관련된 라이브러리는 절대 사용하지 말아야 하며, 이 과제에서는 직접 알고리즘을 구현해야한다.
- 알고리즘이 수렴할 때까지 각 단계를 플롯으로 그린다.예를 들어, 알고리즘이 6번째 단계에서 수렴했다면, 1단계부터 6단계까지의 그림을 제공해야 하며, 각 클러스터는 서로 다른 색상으로 명확하게 구분되어야 한다.
데이터셋
- 2개의 클러스터(K=2)를 사용할 것이며 2D 포인트 데이터는 data_2d.csv 파일을 사용
- 초기 중심점(centroids) 위치는 두 클러스터에 대한 init_centroids.csv 파일에 제공된 위치를 사용할 것이다.
init_centroids.csv
3.323451927212228152e-01,1.798198342198527866e-01
2.241161287440249505e-01,2.219986906322291287e-01
data_2d.csv
0.000000000000000000e+00,-7.687164597386728637e-01,4.608603078297135447e-01
0.000000000000000000e+00,2.687847555392556487e+00,2.366960661575847169e+00
0.000000000000000000e+00,-2.013793555022345139e-01,4.704299346653586511e-01
0.000000000000000000e+00,6.084956800449090597e-01,1.225400029138742575e+00
0.000000000000000000e+00,-8.228190446259109336e-02,1.137218118753473339e+00
0.000000000000000000e+00,2.083069297959621036e+00,2.694482088909215811e+00
0.000000000000000000e+00,1.503019851143946983e+00,1.074847268552238111e+00
0.000000000000000000e+00,3.916620013534907185e-01,-2.874971661363743269e-01
0.000000000000000000e+00,3.213771110785266227e-01,1.296743009602315366e+00
0.000000000000000000e+00,5.912482577647957260e-01,1.267164122169239793e-01
0.000000000000000000e+00,1.150577634973361407e+00,-2.664038442463685374e-01
0.000000000000000000e+00,9.425866685920466503e-01,8.676624226337872337e-01
0.000000000000000000e+00,1.357806126580951567e+00,1.805471547458144421e+00
0.000000000000000000e+00,1.162919909687994968e+00,2.622430134800965540e+00
0.000000000000000000e+00,-9.786851243616156992e-02,1.012305814636828893e+00
0.000000000000000000e+00,8.577741746560831881e-01,1.031965247701047028e+00
0.000000000000000000e+00,6.834367317155296551e-01,1.578139963641977950e-02
0.000000000000000000e+00,1.543771853980922426e+00,1.750230549650776624e+00구현
라이브러리 불러오기
import numpy as np
import matplotlib.pyplot as plt
데이터 불러오기
데이터 포인트 파일 읽기
# 데이터 파일에서 좌표 읽기
def read_data(file_path):
data = []
labels = []
with open(file_path, 'r') as file:
for line in file:
values = line.strip().split(',')
# 첫 번째 값(클러스터 ID)은 무시하고 두 번째, 세 번째 좌표만 사용
data.append([float(values[1]), float(values[2])])
labels.append(int(float(values[0]))) # 초기 클러스터 라벨
return np.array(data), np.array(labels)
- hw2_data_2d.csv 파일에서 데이터를 읽어온다. 각 행은 클러스터 레이블, x 좌표, y 좌표의 형식으로 되어 있다.
- 첫 번째 값(클러스터 레이블)을 제외하고 x, y 좌표만을 데이터로 사용하며, 클러스터 레이블은 따로 저장한다.
중심점 좌표 파일 읽기
[(x1,y1),(x2,y2)] 형태의 numpy 배열로 저장한다.
def read_centroid(file_path):
return np.array(np.loadtxt(file_path, delimiter=',', dtype=float))
Kmeans 클래스 생성
Kmeans 알고리즘 구현을 위한 클래스를 만든다. 다음은 전체 클래스 코드이다.
# K-means Clustering 클래스
class KMeans:
def __init__(self,labels, n_clusters, max_iter=300, tol=1e-5, init_centroids=None):
self.n_clusters = n_clusters # 클러스터 개수 (K=2)
self.max_iter = max_iter # 최대 반복 횟수
self.tol = tol # 수렴 기준 (변화율이 이보다 작아지면 중지)
self.centroids = init_centroids # 초기 클러스터 중심 [(x1,y1),(x2,y2)]
self.labels = labels # 각 좌표가 어떤 클러스터에 속하는지
def fit(self, X):
# 0 단계 시각화 출력
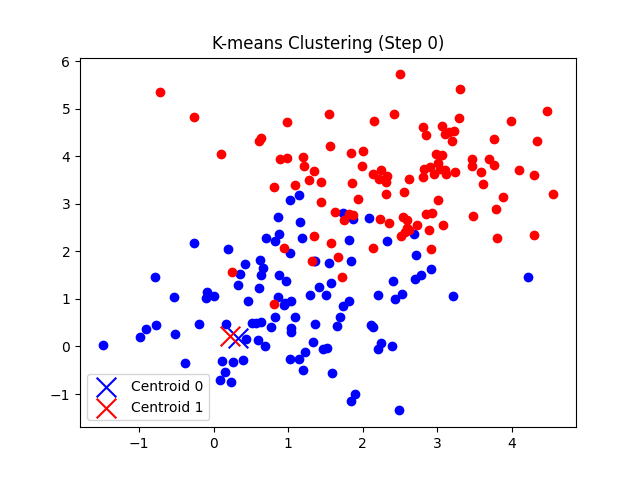
self.plot_step(X,0)
# 초기 중심 설정
for i in range(self.max_iter):
# 각 데이터 포인트에 대해 가장 가까운 클러스터 중심을 업데이트
self.labels = np.array([self.closest_centroid(x) for x in X])
# 이전 중심을 저장 (중심 업데이트 후 비교하기 위해)
old_centroids = self.centroids.copy()
# 각 클러스터마다 해당 클러스터에 속한 좌표 목록을 평균하여 새로운 중심을 계산
for k in range(self.n_clusters):
cluster_points = X[self.labels == k]
if len(cluster_points) > 0:
self.centroids[k] = np.mean(cluster_points, axis=0) # 클러스터 k 의 중심점 좌표 업데이트
# 중심이 얼마나 이동했는지 계산
centroid_shift = np.sum(np.linalg.norm(self.centroids - old_centroids, axis=1))
# 시각화: 각 단계에서의 결과 출력
self.plot_step(X, i + 1)
# 수렴 여부 확인
if centroid_shift < self.tol:
print(f"Converged after {i+1} iterations")
break
# 특정 데이터 포인트에 대해 가장 가까운 클러스터 중심을 찾음
def closest_centroid(self, x):
distances = [euclidean_distance(x, centroid) for centroid in self.centroids]
return np.argmin(distances) # 가장 거리가 가까운 클러스터의 인덱스 반환
# 각 단계별 클러스터링 결과 시각화
def plot_step(self, X, step):
cluster_color_set = ['blue','red']
# 각 클러스터 별 그룹 시각화
for k, centroid in enumerate(self.centroids):
plt.scatter(centroid[0], centroid[1], c=cluster_color_set[k], marker='x', s=200, label=f'Centroid {k}')
plt.scatter(X[self.labels == k ][:,0], X[self.labels == k ][:,1], c=cluster_color_set[k])
plt.title(f"K-means Clustering (Step {step})")
plt.legend()
plt.show()
초기화
def __init__(self,labels, n_clusters, max_iter=300, tol=1e-5, init_centroids=None):
self.n_clusters = n_clusters # 클러스터 개수 (K=2)
self.max_iter = max_iter # 최대 반복 횟수
self.tol = tol # 수렴 기준 (변화율이 이보다 작아지면 중지)
self.centroids = init_centroids # 초기 클러스터 중심 [(x1,y1),(x2,y2)]
self.labels = labels # 각 데이터 포인트가 속하는 클러스터 레이블.
- n_clusters: 클러스터 개수(K), 여기서는 2로 설정.
- max_iter: 최대 반복 횟수. 기본값은 300으로 설정.
- tol: 중심점의 이동이 이 값보다 작으면 수렴했다고 판단하는 기준 (1e-5).
- init_centroids: 초기 중심점의 좌표 (csv 파일에서 불러옴).
- labels: 각 데이터 포인트가 속하는 클러스터 레이블.
모델 학습
주어진 데이터 X에 대해 K-means 알고리즘을 적용하는 메서드이다.
def fit(self, X):
# 0 단계 시각화 출력
self.plot_step(X,0)
# 초기 중심 설정
for i in range(self.max_iter):
# 각 데이터 포인트에 대해 가장 가까운 클러스터 중심을 업데이트
self.labels = np.array([self.closest_centroid(x) for x in X])
# 이전 중심을 저장 (중심 업데이트 후 비교하기 위해)
old_centroids = self.centroids.copy()
# 각 클러스터마다 해당 클러스터에 속한 좌표 목록을 평균하여 새로운 중심을 계산
for k in range(self.n_clusters):
cluster_points = X[self.labels == k]
if len(cluster_points) > 0:
self.centroids[k] = np.mean(cluster_points, axis=0) # 클러스터 k 의 중심점 좌표 업데이트
# 중심이 얼마나 이동했는지 계산
centroid_shift = np.sum(np.linalg.norm(self.centroids - old_centroids, axis=1))
# 시각화: 각 단계에서의 결과 출력
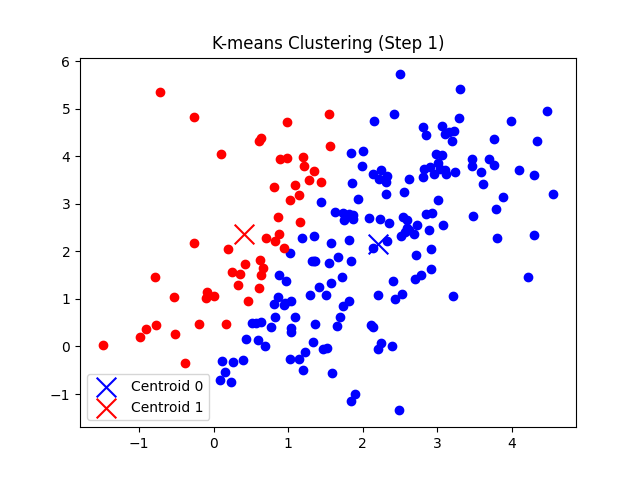
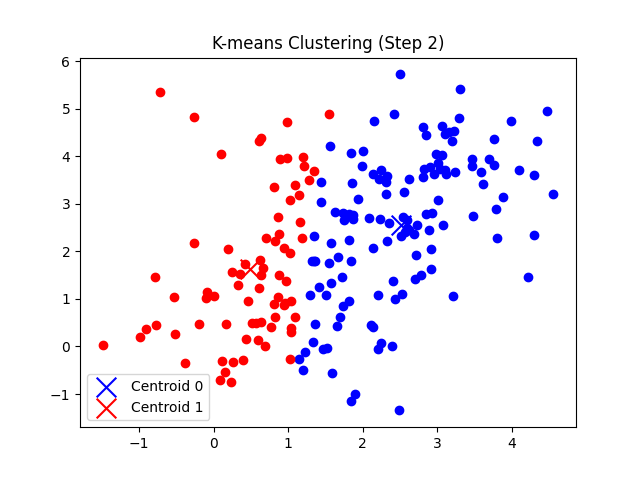
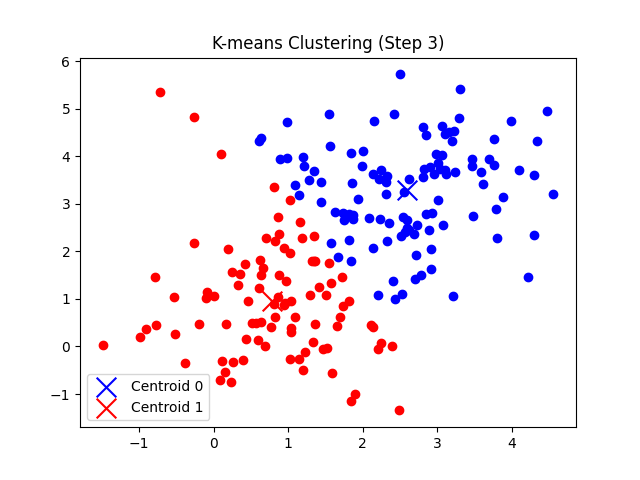
self.plot_step(X, i + 1)
# 수렴 여부 확인
if centroid_shift < self.tol:
print(f"Converged after {i+1} iterations")
break
중심점 초기화 후 반복 과정:
- 0단계에서는 주어진 초기 클러스터를 기준으로 시각화한다.
- 각 데이터 포인트에 대해 가장 가까운 클러스터를 할당하고, 클러스터별로 중심점을 업데이트한다.
- 업데이트 된 이후에 각 단계별로 plot 결과를 출력한다.
- 수렴 여부 확인: 각 반복 단계에서 중심점의 이동량이 tol 값보다 작으면 알고리즘을 종료하고 수렴했다고 판단한다.
이때, 각 데이터 포인트에 대해 가장 가까운 클러스터를 찾기 위해서 거리를 계산할 때 다음의 함수를 이용한다.
각 중심점과의 거리를 계산하여, 가장 가까운 중심점의 인덱스를 반환한다.
# 특정 데이터 포인트에 대해 가장 가까운 클러스터 중심을 찾음
def closest_centroid(self, x):
distances = [euclidean_distance(x, centroid) for centroid in self.centroids]
return np.argmin(distances) # 가장 거리가 가까운 클러스터의 인덱스 반환
클러스터링 결과 시각화
각 반복 단계에서의 클러스터링 결과를 시각화하는 함수이다. 파이플롯 라이브러리를 이용하여 각 좌표가 어떤 클러스터에 속하는지 쉽게 알 수 있도록 색깔을 구분하여 시각화한다.
# 각 단계별 클러스터링 결과 시각화
def plot_step(self, X, step):
cluster_color_set = ['blue','red']
# 각 클러스터 별 그룹 시각화
for k, centroid in enumerate(self.centroids):
plt.scatter(centroid[0], centroid[1], c=cluster_color_set[k], marker='x', s=200, label=f'Centroid {k}')
plt.scatter(X[self.labels == k ][:,0], X[self.labels == k ][:,1], c=cluster_color_set[k])
plt.title(f"K-means Clustering (Step {step})")
plt.legend()
plt.show()실행
데이터 파일을 로드하고, K-means 알고리즘을 실행한다.
def main():
# 데이터 파일 로드
data_file = './data/hw2_data_2d.csv'
centroids_file = './data/hw2_2d_init_centroids.csv'
X, labels = read_data(data_file) # 데이터 포인트 로드
init_centroids = read_centroid(centroids_file) # 초기 중심 로드
# K-means 클러스터링
kmeans = KMeans(labels=labels ,n_clusters=2, init_centroids=init_centroids)
kmeans.fit(X)
if __name__ == '__main__':
main()결과
총 7번의 반복 후에 클러스터의 중심점 좌표가 tol(1e-5) 미만으로 움직이게 되므로 클러스터링이 완료되었다고 판단한다.
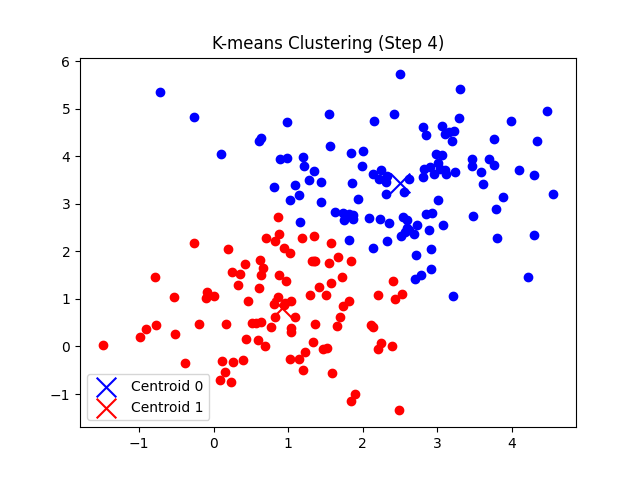
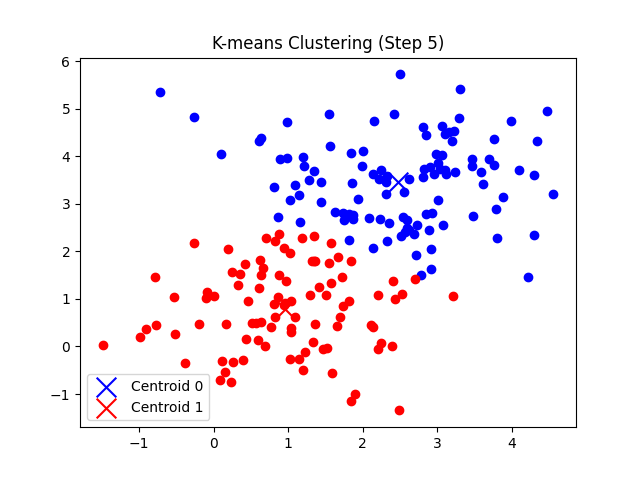
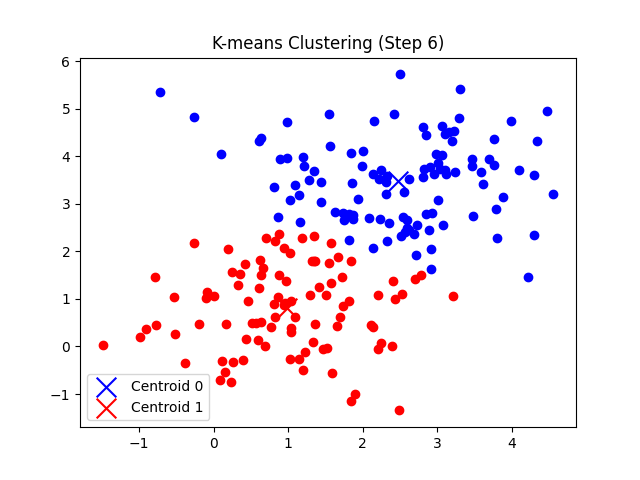
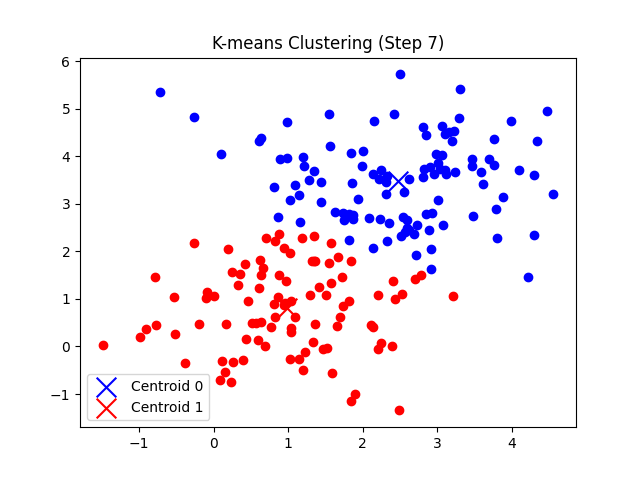
'CS > 인공지능' 카테고리의 다른 글
| [24-2] 👾 기계학습(ML) 프로젝트 : 외계 행성 찾기 👽 (0) | 2024.11.25 |
|---|---|
| SVM 을 활용한 스팸 분류기 ( Spam Classification via SVM ) (1) | 2024.10.17 |
| 나이브 베이즈를 사용한 스팸 메일 분류기 (Spam Classification via Naïve Bayes) (3) | 2024.10.17 |
| [Tensorflow keras] image generation using Stable Diffusion (0) | 2024.06.24 |
| Simple Diffusion Image generate Model (0) | 2024.06.21 |



